Asgard (archaea)
Asgard or Asgardarchaeota[2] is a proposed superphylum consisting of a group of archaea that includes Lokiarchaeota, Thorarchaeota, Odinarchaeota, and Heimdallarchaeota.[3] A representative of the group was cultivated.[4] The Asgard superphylum represents the closest prokaryotic relatives of eukaryotes,[5] which possibly emerged from an ancestral lineage of Asgardarchaeota after assimilating bacteria through the process of symbiogenesis.[5][6]
| Asgard | |
|---|---|
| Scientific classification | |
| Domain: | Archaea |
| Kingdom: | Proteoarchaeota |
| Superphylum: | Asgard (archaea) Katarzyna Zaremba-Niedzwiedzka, et al. 2017 |
| Phyla | |
| |
 | |
| Synonyms[1] | |
| |
Discovery
In the summer of 2010, sediments from a gravity core taken in the rift valley on the Knipovich ridge in the Arctic Ocean, near the so-called Loki's Castle hydrothermal vent site, were analysed. Specific sediment horizons previously shown to contain high abundances of novel archaeal lineages, were subjected to metagenomic analysis.[7][8]
In 2015, an Uppsala University-led team proposed the Lokiarchaeota phylum based on phylogenetic analyses using a set of highly conserved protein-coding genes.[9] Through a reference to the hydrothermal vent complex from which the first genome sample originated, the name refers to Loki, the Norse shape-shifting god.[10] The Loki of mythology has been described as "a staggeringly complex, confusing, and ambivalent figure who has been the catalyst of countless unresolved scholarly controversies",[11] analogous to the role of Lokiarchaeota in the debates about the origin of eukaryotes.[9][12]
In 2016, a University of Texas-led team discovered Thorarchaeota from samples taken from the White Oak River in North Carolina, named in reference to Thor, another Norse god.[13]
Additional samples from Loki's Castle, Yellowstone National Park, Aarhus Bay, an aquifer near the Colorado River, New Zealand's Radiata Pool, hydrothermal vents near Taketomi Island, Japan, and the White Oak River estuary in the United States led researchers to discover Odinarchaeota and Heimdallarchaeota,[3] and following the naming convention having been established to use Norse deities, the archaea were named for Odin and Heimdallr, respectively. Researchers therefore, named the superphylum containing these microbes “Asgard”, after the realm of the deities in Norse mythology.[3]
Description
Asgard members encode many eukaryotic signature proteins, including novel GTPases, membrane-remodelling proteins like ESCRT and SNF7, a ubiquitin modifier system, and N-glycosylation pathway homologs.[3]
Asgard archaeons have a regulated actin cytoskeleton, and the profilins and gelsolins they use can interact with eukaryotic actins.[14][15][16] They also seem to form vesicles under Cryogenic electron microscopy. Some may have a PKD domain S-layer.[4] They also share the three-way ES39 expansion in LSU rRNA with eukaryotes.[17]
Metabolism
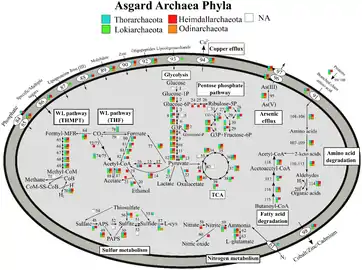 Metabolic pathways of Asgard archaea, variation by Phyla[18]
Metabolic pathways of Asgard archaea, variation by Phyla[18]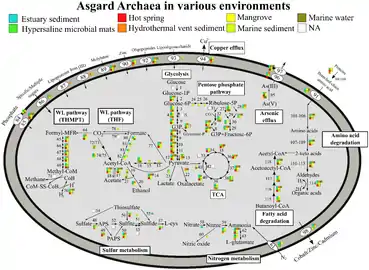 Metabolic pathways of Asgard archaea, variation by environment[18]
Metabolic pathways of Asgard archaea, variation by environment[18]
Asgard archaea are obligate anaerobes. They have a Wood–Ljungdahl pathway and perform glycolysis. Members can be autotrophs, heterotrophs, or phototrophs using heliorhodopsin.[18] One member, Candidatus Prometheoarchaeum syntrophicum, performs syntrophy with a sulfur-reducing proteobacteria and a methanogenic archaea.[4]
The RuBisCO they have are not carbon-fixing, but likely used for nucleoside salvaging.[18]
Eukaryotic-like features in subdivisions
The phylum "Heimdallarchaeota" was found to have N-terminal core histone tails, a feature previously thought to be exclusively eukaryotic, in 2017. Two other archaeal phyla, both outside of Asgard, were found to also have tails in 2018.[19]
In January 2020, scientists found Candidatus Prometheoarchaeum syntrophicum, a member of Lokiarcheota, engaging in cross-feeding with two bacterial species. Drawing an analogy to symbiogenesis, they consider this relationship a possible link between the simple prokaryotic microorganisms and the complex eukaryotic microorganisms occurring approximately two billion years ago.[20][4]
Classification
The phylogenetic relationship of this group is still under discussion. The relationship of the members is approximately as follows:[5][6]
| Proteoarchaeota |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The Heimdallarchaeota are considered the deepest branching Asgard archaea.[4] The Eukaryotes may be sister to the Heimdallarchaeota or the Asgard archaea. A favored scenario is syntrophy, where one organism depends on the feeding of the other. In this case, the syntrophy may have been due to the Asgard archaea having been incorporated in an unknown type of bacteria, developing into the nucleus. An α-proteobacterium was incorporated to become the mitochondrion.[21]
References
- Fournier GP, Poole AM. (2018). "A Briefly Argued Case That Asgard Archaea Are Part of the Eukaryote Tree". Front. Microbiol. 9: 1896. doi:10.3389/fmicb.2018.01896. PMC 6104171. PMID 30158917.CS1 maint: uses authors parameter (link)
- Violette Da Cunha, Morgan Gaia, Daniele Gadelle, Arshan Nasir, Patrick Forterre: Lokiarchaea are close relatives of Euryarchaeota, not bridging the gap between prokaryotes and eukaryotes, in: PLoS Genet. 2017 Jun; 13(6): e1006810. 2017 Jun 12, doi: 10.1371/journal.pgen.1006810
- Zaremba-Niedzwiedzka, Katarzyna; Caceres, Eva F.; Saw, Jimmy H.; Bäckström, Disa; Juzokaite, Lina; Vancaester, Emmelien; Seitz, Kiley W.; Anantharaman, Karthik; Starnawski, Piotr (11 January 2017). "Asgard archaea illuminate the origin of eukaryotic cellular complexity" (PDF). Nature. 541 (7637): 353–358. Bibcode:2017Natur.541..353Z. doi:10.1038/nature21031. ISSN 1476-4687. PMID 28077874.
- Imachi, Hiroyuki; Nobu, Masaru K.; Nakahara, Nozomi; Morono, Yuki; Ogawara, Miyuki; Takaki, Yoshihiro; Takano, Yoshinori; Uematsu, Katsuyuki; Ikuta, Tetsuro; Ito, Motoo; Matsui, Yohei (2020-01-23). "Isolation of an archaeon at the prokaryote–eukaryote interface". Nature. 577 (7791): 519–525. Bibcode:2020Natur.577..519I. doi:10.1038/s41586-019-1916-6. ISSN 1476-4687. PMC 7015854. PMID 31942073.
- Eme, Laura; Spang, Anja; Lombard, Jonathan; Stairs, Courtney W.; Ettema, Thijs J. G. (10 November 2017). "Archaea and the origin of eukaryotes". Nature Reviews Microbiology. 15 (12): 711–723. doi:10.1038/nrmicro.2017.133. ISSN 1740-1534. PMID 29123225.
- Williams, Tom A.; Cox, Cymon J.; Foster, Peter G.; Szöllősi, Gergely J.; Embley, T. Martin (2019-12-09). "Phylogenomics provides robust support for a two-domains tree of life". Nature Ecology & Evolution. 4 (1): 138–147. doi:10.1038/s41559-019-1040-x. ISSN 2397-334X. PMC 6942926. PMID 31819234.
- Jørgensen, Steffen Leth; Hannisdal, Bjarte; Lanzen, Anders; Baumberger, Tamara; Flesland, Kristin; Fonseca, Rita; Øvreås, Lise; Steen, Ida H; Thorseth, Ingunn H; Pedersen, Rolf B; Schleper, Christa (September 5, 2012). "Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge". PNAS. 109 (42): E2846–55. doi:10.1073/pnas.1207574109. PMC 3479504. PMID 23027979.
- Jørgensen, Steffen Leth; Thorseth, Ingunn H; Pedersen, Rolf B; Baumberger, Tamara; Schleper, Christa (October 4, 2013). "Quantitative and phylogenetic study of the Deep Sea Archaeal Group in sediments of the Arctic mid-ocean spreading ridge". Frontiers in Microbiology. 4: 299. doi:10.3389/fmicb.2013.00299. PMC 3790079. PMID 24109477.
- Spang, Anja; Saw, Jimmy H.; Jørgensen, Steffen L.; Zaremba-Niedzwiedzka, Katarzyna; Martijn, Joran; Lind, Anders E.; Eijk, Roel van; Schleper, Christa; Guy, Lionel (May 2015). "Complex archaea that bridge the gap between prokaryotes and eukaryotes". Nature. 521 (7551): 173–179. Bibcode:2015Natur.521..173S. doi:10.1038/nature14447. ISSN 1476-4687. PMC 4444528. PMID 25945739.
- Yong, Ed. "Break in the Search for the Origin of Complex Life". The Atlantic. Retrieved 2018-03-21.
- von Schnurbein, Stefanie (November 2000). "The Function of Loki in Snorri Sturluson's "Edda"". History of Religions. 40 (2): 109–124. doi:10.1086/463618.
- Spang, Anja; Eme, Laura; Saw, Jimmy H.; Caceres, Eva F.; Zaremba-Niedzwiedzka, Katarzyna; Lombard, Jonathan; Guy, Lionel; Ettema, Thijs J. G.; Rokas, Antonis (29 March 2018). "Asgard archaea are the closest prokaryotic relatives of eukaryotes". PLOS Genetics. 14 (3): e1007080. doi:10.1371/journal.pgen.1007080. PMC 5875740. PMID 29596421.
- Seitz, Kiley W; Lazar, Cassandre S; Hinrichs, Kai-Uwe; Teske, Andreas P; Baker, Brett J (29 January 2016). "Genomic reconstruction of a novel, deeply branched sediment archaeal phylum with pathways for acetogenesis and sulfur reduction". The ISME Journal. 10 (7): 1696–1705. doi:10.1038/ismej.2015.233. ISSN 1751-7370. PMC 4918440. PMID 26824177.
- Akıl, Caner; Robinson, Robert C. (3 October 2018). "Genomes of Asgard archaea encode profilins that regulate actin". Nature. 562 (7727): 439–443. Bibcode:2018Natur.562..439A. doi:10.1038/s41586-018-0548-6. PMID 30283132.
- Akıl, Caner; Tran, Linh T.; Orhant-Prioux, Magali; Baskaran, Yohendran; Manser, Edward; Blanchoin, Laurent; Robinson, Robert C. (14 September 2019). "Complex eukaryotic-like actin regulation systems from Asgard archaea". bioRxiv 10.1101/768580.
- Akıl, Caner; Tran, Linh T.; Orhant-Prioux, Magali; Baskaran, Yohendran; Manser, Edward; Blanchoin, Laurent; Robinson, Robert C. (18 August 2020). "Insights into the evolution of regulated actin dynamics via characterization of primitive gelsolin/cofilin proteins from Asgard archaea". PNAS. 117 (33): 19904–19913. doi:10.1073/pnas.2009167117. PMID 32747565.
- Penev, Petar I; Fakhretaha-Aval, Sara; Patel, Vaishnavi J; Cannone, Jamie J; Gutell, Robin R; Petrov, Anton S; Williams, Loren Dean; Glass, Jennifer B (1 October 2020). "Supersized Ribosomal RNA Expansion Segments in Asgard Archaea". Genome Biology and Evolution. 12 (10): 1694–1710. doi:10.1093/gbe/evaa170.
- MacLeod, Fraser; Kindler, Gareth S.; Wong, Hon Lun; Chen, Ray; Burns, Brendan P. (2019). "Asgard archaea: Diversity, function, and evolutionary implications in a range of microbiomes". AIMS Microbiology. 5 (1): 48–61. doi:10.3934/microbiol.2019.1.48. ISSN 2471-1888. PMC 6646929. PMID 31384702.
- Henneman B, van Emmerik C, van Ingen H, Dame RT (September 2018). "Structure and function of archaeal histones". PLOS Genetics. 14 (9): e1007582. Bibcode:2018BpJ...114..446H. doi:10.1371/journal.pgen.1007582. PMC 6136690. PMID 30212449.
- Zimmer, Carl (15 January 2020). "This Strange Microbe May Mark One of Life's Great Leaps - A organism living in ocean muck offers clues to the origins of the complex cells of all animals and plants". The New York Times. Retrieved 16 January 2020.
- López-García, Purificación; Moreira, David (2019-07-01). "Eukaryogenesis, a syntrophy affair". Nature Microbiology. 4 (7): 1068–1070. doi:10.1038/s41564-019-0495-5. ISSN 2058-5276. PMC 6684364. PMID 31222170.
External links
- Traci Watson: The trickster microbes that are shaking up the tree of life, in: Nature, 14 May 2019