Retinal dehydrogenase
In enzymology, a retinal dehydrogenase, also known as retinaldehyde dehydrogenase, catalyzes the chemical reaction converting retinal to retinoic acid. This enzyme belongs to the family of oxidoreductases, specifically the class acting on aldehyde or oxo- donor groups with NAD+ or NADP+ as acceptor groups, the systematic name being retinal:NAD+ oxidoreductase. This enzyme participates in retinol metabolism. The general scheme for the reaction catalyzed by this enzyme is:
| retinal dehydrogenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 retinal dehydrogenase inhibited by Yb | |||||||||
| Identifiers | |||||||||
| EC number | 1.2.1.36 | ||||||||
| CAS number | 37250-99-0 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
retinal + NAD+ + H2O retinoic acid + NADH + H+
Structure
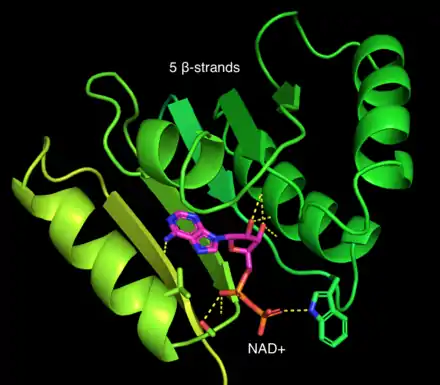
Retinal dehydrogenase is a tetramer of identical units, consisting of a dimer of dimers.[1] Retinal dehydrogenase monomers are composed of three domains: a nucleotide-binding domain, a tetramerization domain, and a catalytic domain. The dimer can be pictured as an "X" with the dimers forming upper and lower halves that cross over each other. Interestingly, the nucleotide-binding domain of retinal dehydrogenase contains 5 instead of the usual 6 β-strands in the Rossman fold.[2] This appears to be conserved across many aldehyde dehydrogenases. The tetramerization domains lie equatorially along the "X" and the nucleotide binding regions appear on the tips of the "X". Nearby the tetramerization domain lies a 12 Å deep tunnel that gives the substrate access to the key catalytic regions.[1] Residues near the C-terminal end of the catalytic domain have been found to impart specificity in other aldehyde dehydrogenases. Common to many aldehyde dehydrogenases is a catalytic cysteine, which was found to be present in RALDH2, a specific retinal dehydrogenase for which the structure has been solved.[1][3][4]
Specificity
There are three general classes of aldehyde dehydrogenases: class 1 (ALDH1) comprises cytosolic proteins, class 2 (ALDH2) includes mitochondrial proteins, and class 3 (ALDH3) includes tumor-related proteins.[4] ALDH1 enzymes show a high specificity for all-trans retinal and 9-cis retinal in kinetic studies of sheep liver aldehyde dehydrogenases while ALDH2 enzymes show little affinity for retinal and instead appears to be mainly involved in the oxidation of acetaldehyde.[5][6] The entrance tunnel to the enzyme active site appears to provide the specificity observed in ALDH1 for retinal as a substrate. The size of the tunnel is key in imparting this specificity: the solvent-accessible diameter of the entrance tunnel is 150 Å3 in ALDH1, so the relatively large retinal can be accommodated while the solvent accessible diameter in ALDH2 is only 20 Å3 which limits accessibility to retinal but amply accommodates acetaldehyde.[7]
Mechanism
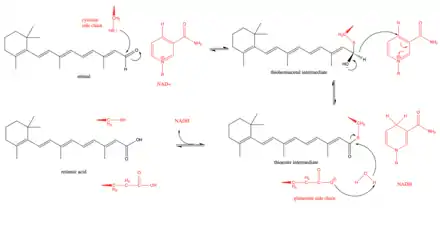
The proposed mechanism of retinal dehydrogenase begins with a key cysteine residue in the active site attacking the aldehyde group in retinal to form a thiohemiacetal intermediate.[3] Then, a hydride shift is facilitated by the enzyme to form NADH and a thioester intermediate. This hydride shift has been shown to be stereospecific in a subset (class 3) of retinal dehydrogenases.[8] The thioester intermediate is then attacked by a water molecule, which is made more nucleophilic by a glutamate residue that lies near the active site.[9] There has been some debate as to whether the glutamate residue near the active site acts as a general base during the reaction or whether it is more limited and merely deprotonates the catalytic cysteine to make the cysteine more nucleophilic.[9] Kinetic studies have supported this mechanism by showing that the reaction follows an ordered sequential path with NAD+ binding first which is followed by the binding of retinal, the catalytic breakdown of retinal to retinoic acid, the release of retinoic acid, and finally the release of NADH.[10]
Regulation
Some of the strategies for regulating retinal dehydrogenases are only now becoming more clear after in vivo regulation remained mysterious for some time, though much of the current research on regulation has focused on the modulation of gene expression rather than direct protein regulation.[7] Dendritic cells in the gut help in modulating immune tolerance through the activity of retinal dehydrogenase; expression in these cells may be driven by a TNF receptor, 4-1-BB.[11] It was also shown that the expression of a certain retinal dehydrogenase found in humans, retinal short-chain dehydrogenase/reductase (retSDR1), is increased by tumor-suppressor proteins p53 and p63, suggesting that retSDR1 may have tumor-preventing activities.[12] Expression of retinal dehydrogenase types 1 and 2 genes is enhanced by the addition of cholesterol or cholesterol derivatives.[13] Disulfiram is a drug used to artificially regulate aldehyde dehydrogenase activity in patients with alcoholism by inhibiting the activity of aldehyde dehydrogenases, though it is not specific to retinal dehydrogenase.[14] Other exogenous molecules have also been found to inhibit retinal dehydrogenase activity including nitrofen, 4-biphenyl carboxylic acid, bisdiamine, and SB-210661.[15]
Clinical significance
Retinal dehydrogenase plays a key role in the biosynthesis of retinoic acid, which in turn acts in cell signaling pathways. Retinoic acid is distinct from other cell signaling molecules in that it diffuses into the nucleus and binds directly to gene targets via retinoic acid receptors.[16] This retinoic acid signaling pathway also appears to be unique to chordates, as suggested by the presence of retinal dehydrogenases exclusively in chordates.[17] Retinoic acid signaling appears to control developmental processes like neurogenesis, cardiogenesis, forelimb bud development, foregut development, and eye development. Retinoic acid signaling is also important for maintaining adult neuronal and epithelium cell type.[18] Retinoic acid is generated in organisms by first oxidizing retinol (Vitamin A) to retinal with an alcohol dehydrogenase. Then, a retinal dehydrogenase oxidizes retinal to retinoic acid. The production of retinoic acid from vitamin A must be tightly controlled as high levels of retinoic acid and vitamin A can lead to toxic effects, while vitamin A deficiency leads to its own issues in development.[19][20] This provides a rationale for many of the transcriptional regulatory strategies discussed earlier. In humans, mutations in a gene coding for a certain retinal dehydrogenase (RDH12) can also lead to Leber's congenital amaurosis, a retinal dystrophy responsible for many cases of congenital blindness.[21]
Isoforms
Different isoforms of retinal dehydrogenase exist and play a key role in development, as the types are differentially expressed inside a developing embryo. The enzyme retinal dehydrogenase type-2 (RALDH2) catalyzes much of the retinoic acid formation during development, but not all. RALDH2 is crucial for development midgestation and helps drive neural, heart, lung, and forelimb development; it is also responsible for all retinoic acid development during certain periods of midgestation.[22] Later in development, retinal dehydrogenase type-1 (RALDH1) begins activity in the dorsal pit of the retina and retinal dehydrogenase type-3 (RALDH3) becomes active in the olfactory pit, ventral retina, and urinary tract. Raldh2 gene knockouts are fatal in mice during development since the brain cannot develop normally.[23] Raldh3 gene knockout is fatal at birth in mice since nasal passages are not properly developed and instead are blocked.[24] Raldh1 knockouts are not fatal and, interestingly, have been shown to be protective against diet-induced obesity in mice in a retinoid-independent manner.[25]
References
- Lamb AL, Newcomer ME (May 1999). "The structure of retinal dehydrogenase type II at 2.7 A resolution: implications for retinal specificity". Biochemistry. 38 (19): 6003–11. doi:10.1021/bi9900471. PMID 10320326.
- Liu ZJ, Sun YJ, Rose J, Chung YJ, Hsiao CD, Chang WR, Kuo I, Perozich J, Lindahl R, Hempel J, Wang BC (April 1997). "The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold". Nature Structural Biology. 4 (4): 317–26. doi:10.1038/nsb0497-317. PMID 9095201.
- Abriola DP, Fields R, Stein S, MacKerell AD, Pietruszko R (September 1987). "Active site of human liver aldehyde dehydrogenase". Biochemistry. 26 (18): 5679–84. doi:10.1021/bi00392a015. PMID 3676276.
- Farrés J, Wang TT, Cunningham SJ, Weiner H (February 1995). "Investigation of the active site cysteine residue of rat liver mitochondrial aldehyde dehydrogenase by site-directed mutagenesis". Biochemistry. 34 (8): 2592–8. doi:10.1021/bi00008a025. PMID 7873540.
- Yoshida A, Hsu LC, Davé V (1992). "Retinal oxidation activity and biological role of human cytosolic aldehyde dehydrogenase". Enzyme. 46 (4–5): 239–44. doi:10.1159/000468794. PMID 1292933.
- Kitson KE, Blythe TJ (1999). "The hunt for a retinal-specific aldehyde dehydrogenase in sheep liver". Advances in Experimental Medicine and Biology. 463: 213–21. doi:10.1007/978-1-4615-4735-8_26. ISBN 978-1-4613-7146-5. PMID 10352688.
- Moore SA, Baker HM, Blythe TJ, Kitson KE, Kitson TM, Baker EN (December 1998). "Sheep liver cytosolic aldehyde dehydrogenase: the structure reveals the basis for the retinal specificity of class 1 aldehyde dehydrogenases". Structure. 6 (12): 1541–51. doi:10.1016/S0969-2126(98)00152-X. PMID 9862807.
- Jones KH, Lindahl R, Baker DC, Timkovich R (August 1987). "Hydride transfer stereospecificity of rat liver aldehyde dehydrogenases". The Journal of Biological Chemistry. 262 (23): 10911–3. PMID 3038902.
- Wang X, Weiner H (January 1995). "Involvement of glutamate 268 in the active site of human liver mitochondrial (class 2) aldehyde dehydrogenase as probed by site-directed mutagenesis". Biochemistry. 34 (1): 237–43. doi:10.1021/bi00001a028. PMID 7819202.
- Hart GJ, Dickinson FM (June 1982). "Kinetic properties of highly purified preparations of sheep liver cytoplasmic aldehyde dehydrogenase". The Biochemical Journal. 203 (3): 617–27. doi:10.1042/bj2030617. PMC 1158276. PMID 7115304.
- Lee SW, Park Y, Eun SY, Madireddi S, Cheroutre H, Croft M (September 2012). "Cutting edge: 4-1BB controls regulatory activity in dendritic cells through promoting optimal expression of retinal dehydrogenase". Journal of Immunology. 189 (6): 2697–701. doi:10.4049/jimmunol.1201248. PMC 3436963. PMID 22896640.
- Kirschner RD, Rother K, Müller GA, Engeland K (June 2010). "The retinal dehydrogenase/reductase retSDR1/DHRS3 gene is activated by p53 and p63 but not by mutants derived from tumors or EEC/ADULT malformation syndromes". Cell Cycle. 9 (11): 2177–88. doi:10.4161/cc.9.11.11844. PMID 20543567.
- Huq MD, Tsai NP, Gupta P, Wei LN (July 2006). "Regulation of retinal dehydrogenases and retinoic acid synthesis by cholesterol metabolites". The EMBO Journal. 25 (13): 3203–13. doi:10.1038/sj.emboj.7601181. PMC 1500992. PMID 16763553.
- Lipsky JJ, Berti JJ, Aquilina JW, Mays DC (October 1997). "Effect of a disulfiram metabolite on retinaldehyde metabolism". Lancet. 350 (9085): 1176. doi:10.1016/S0140-6736(05)63821-4. PMID 9343525.
- Mey J, Babiuk RP, Clugston R, Zhang W, Greer JJ (February 2003). "Retinal dehydrogenase-2 is inhibited by compounds that induce congenital diaphragmatic hernias in rodents". The American Journal of Pathology. 162 (2): 673–9. doi:10.1016/S0002-9440(10)63861-8. PMC 1851155. PMID 12547725.
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ (November 2001). "Nuclear receptors and lipid physiology: opening the X-files". Science. 294 (5548): 1866–70. Bibcode:2001Sci...294.1866C. doi:10.1126/science.294.5548.1866. PMID 11729302.
- Marlétaz F, Holland LZ, Laudet V, Schubert M (2006). "Retinoic acid signaling and the evolution of chordates". International Journal of Biological Sciences. 2 (2): 38–47. doi:10.7150/ijbs.2.38. PMC 1458431. PMID 16733532.
- Maden M (October 2007). "Retinoic acid in the development, regeneration and maintenance of the nervous system". Nature Reviews. Neuroscience. 8 (10): 755–65. doi:10.1038/nrn2212. PMID 17882253.
- Guillonneau M, Jacqz-Aigrain E (September 1997). "[Teratogenic effects of vitamin A and its derivates]". Archives de Pédiatrie. 4 (9): 867–74. doi:10.1016/S0929-693X(97)88158-4. PMID 9345570.
- Dickman ED, Thaller C, Smith SM (August 1997). "Temporally-regulated retinoic acid depletion produces specific neural crest, ocular and nervous system defects". Development. 124 (16): 3111–21. PMID 9272952.
- Perrault I, Hanein S, Gerber S, Barbet F, Ducroq D, Dollfus H, Hamel C, Dufier JL, Munnich A, Kaplan J, Rozet JM (October 2004). "Retinal dehydrogenase 12 (RDH12) mutations in leber congenital amaurosis". American Journal of Human Genetics. 75 (4): 639–46. doi:10.1086/424889. PMC 1182050. PMID 15322982.
- Molotkova N, Molotkov A, Sirbu IO, Duester G (February 2005). "Requirement of mesodermal retinoic acid generated by Raldh2 for posterior neural transformation". Mechanisms of Development. 122 (2): 145–55. doi:10.1016/j.mod.2004.10.008. PMC 2826194. PMID 15652703.
- Mic FA, Haselbeck RJ, Cuenca AE, Duester G (May 2002). "Novel retinoic acid generating activities in the neural tube and heart identified by conditional rescue of Raldh2 null mutant mice". Development. 129 (9): 2271–82. PMC 2833017. PMID 11959834.
- Molotkov A, Molotkova N, Duester G (May 2006). "Retinoic acid guides eye morphogenetic movements via paracrine signaling but is unnecessary for retinal dorsoventral patterning". Development. 133 (10): 1901–10. doi:10.1242/dev.02328. PMC 2833011. PMID 16611695.
- Yang D, Krois CR, Huang P, Wang J, Min J, Yoo HS, Deng Y, Napoli JL (November 2, 2017). "Raldh1 promotes adiposity during adolescence independently of retinal signaling". PLOS ONE. 12 (11): e0187669. Bibcode:2017PLoSO..1287669Y. doi:10.1371/journal.pone.0187669. PMC 5667840. PMID 29095919.