Prime editing
Prime editing is a ‘search-and-replace’ genome editing technology in molecular biology by which the genome of living organisms may be modified. The technology directly writes new genetic information into a targeted DNA site. It uses a fusion protein, consisting of a catalytically impaired Cas9 endonuclease fused to an engineered reverse transcriptase enzyme, and a prime editing guide RNA (pegRNA), capable of identifying the target site and providing the new genetic information to replace the target DNA nucleotides. It mediates targeted insertions, deletions, and base-to-base conversions without the need for double strand breaks (DSBs) or donor DNA templates.[1]
The technology is an early-stage, experimental genome editing method that has received mainstream press attention due to its potential uses in medical genetics. It utilizes similar methodologies to precursor genome editing technologies, including CRISPR/Cas9 and base editors. As of 2019, it remains a scientific proof of concept, with no therapeutic applications.[1]
Genome editing
Components
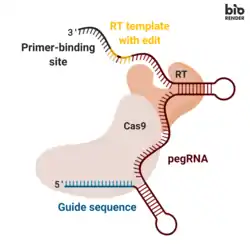
Prime editing involves three major components:[1]
- A prime editing guide RNA (pegRNA), capable of (i) identifying the target nucleotide sequence to be edited, and (ii) encoding new genetic information that replaces the targeted sequence. The pegRNA consists of an extended single guide RNA (sgRNA) containing a primer binding site (PBS) and a reverse transcriptase (RT) template sequence. During genome editing, the primer binding site allows the 3’ end of the nicked DNA strand to hybridize to the pegRNA, while the RT template serves as a template for the synthesis of edited genetic information.[1]
- A fusion protein consisting of a Cas9 H840A nickase fused to a Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase.[1]
- Cas9 H840A nickase: the Cas9 enzyme contains two nuclease domains that can cleave DNA sequences, a RuvC domain that cleaves the non-target strand and a HNH domain that cleaves the target strand. The introduction of a H840A substitution in Cas9, through which the 840º amino acid histidine is replaced by an alanine, inactivates the HNH domain. With only the RuvC functioning domain, the catalytically impaired Cas9 introduces a single strand nick, hence the name nickase.[2]
- M-MLV reverse transcriptase: an enzyme that synthesizes DNA from a single-stranded RNA template.[1]
- A single guide RNA (sgRNA) that directs the Cas9 H840A nickase portion of the fusion protein to nick the non-edited DNA strand.[1]
Mechanism

Genomic editing takes place by transfecting cells with the pegRNA and the fusion protein. Transfection is often accomplished by introducing vectors into a cell. Once internalized, the fusion protein nicks the target DNA sequence, exposing a 3’-hydroxyl group that can be used to initiate (prime) the reverse transcription of the RT template portion of the pegRNA. This results in a branched intermediate that contains two DNA flaps: a 3’ flap that contains the newly synthesized (edited) sequence, and a 5’ flap that contains the dispensable, unedited DNA sequence. The 5’ flap is then cleaved by structure-specific endonucleases or 5’ exonucleases. This process allows 3’ flap ligation, and creates a heteroduplex DNA composed of one edited strand and one unedited strand. The reannealed double stranded DNA contains nucleotide mismatches at the location where editing took place. In order to correct the mismatches, the cells exploit the intrinsic mismatch repair mechanism, with two possible outcomes: (i) the information in the edited strand is copied into the complementary strand, permanently installing the edit; (ii) the original nucleotides are re-incorporated into the edited strand, excluding the edit.[1]
Development process
During the development of this technology, several modifications were done to the components, in order to increase its effectiveness.[1]
Prime editor 1
In the first system, a wild-type Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase was fused to the Cas9 H840A nickase C-terminus. Detectable editing efficiencies were observed.[1]
Prime editor 2
In order to enhance DNA-RNA affinity, enzyme processivity, and thermostability, five amino acid substitutions were incorporated into the M-MLV reverse transcriptase. The mutant M-MLV RT was then incorporated into PE1 to give rise to (Cas9 (H840A)-M-MLV RT(D200N/L603W/T330P/T306K/W313F)). Efficiency improvement was observed over PE1.[1]

Prime editor 3
Despite its increased efficacy, the edit inserted by PE2 might still be removed due to DNA mismatch repair of the edited strand. To avoid this problem during DNA heteroduplex resolution, an additional single guide RNA (sgRNA) is introduced. This sgRNA is designed to match the edited sequence introduced by the pegRNA, but not the original allele. It directs the Cas9 nickase portion of the fusion protein to nick the unedited strand at a nearby site, opposite to the original nick. Nicking the non-edited strand causes the cell’s natural repair system to copy the information in the edited strand to the complementary strand, permanently installing the edit.[1]
Implications
Although additional research is required to improve the efficiency of prime editing, the technology offers promising scientific improvements over other gene editing tools. The prime editing technology has the potential to correct the vast majority of pathogenic alleles that cause genetic diseases, as it can repair insertions, deletions, and nucleotide substitutions.[1]
Advantages
The prime editing tool offers advantages over traditional gene editing technologies. CRISPR/Cas9 edits rely on non-homologous end joining (NHEJ) or homology-directed repair (HDR) to fix DNA breaks, while the prime editing system employs DNA mismatch repair. This is an important feature of this technology given that DNA repair mechanisms such as NHEJ and HDR, generate unwanted, random insertions or deletions (INDELs) byproducts which complicate the retrieval of cells carrying the correct edit.[1][3]
The prime system introduces single-stranded DNA breaks instead of the double-stranded DNA breaks observed in other editing tools, such as base editors. Collectively, base editing and prime editing offer complementary strengths and weaknesses for making targeted transition mutations. Base editors offer higher editing efficiency and fewer INDEL byproducts if the desired edit is a transition point mutation and a PAM sequence exists roughly 15 bases from the target site. However, because the prime editing technology does require a precisely positioned PAM sequence to target a nucleotide sequence, it offers more flexibility and editing precision. Remarkably, prime editors allow all types of substitutions, transitions and transversions to be inserted into the target sequence.[1][3]
Because the prime system involves three separate DNA binding events (between (i) the guide sequence and the target DNA, (ii) the primer binding site and the target DNA, and (iii) the 3’ end of the nicked DNA strand and the pegRNA), it has been suggested to have fewer undesirable off-target effects than CRISPR/Cas9.[1][3]
Limitations
There is considerable interest in applying gene editing methods to the treatment of diseases with a genetic component. However, there are multiple challenges associated with this approach. An effective treatment would require editing of a large number of target cells, which in turn would require an effective method of delivery and a great level of tissue specificity.[1][4]
As of 2019, prime editing looks promising for relatively small genetic alterations, but more research needs to be conducted to evaluate whether the technology is efficient in making larger alterations, such as targeted insertions and deletions. Larger genetic alterations would require a longer RT template, which could hinder the efficient delivery of pegRNA to target cells. Furthermore, a pegRNA containing a long RT template could become vulnerable to damage caused by cellular enzymes.[1][4]
Overall, much research will be needed before prime editing could be used to correct pathogenic alleles in human diseases.[1][4]
Delivery method
Base editors used for prime editing require delivery of both a protein and RNA molecule into living cells. Introducing exogenous gene editing technologies into living organisms is a significant challenge. One potential way to introduce a base editor into animals and plants would be to package the base editor into a viral capsid. The target organism can then be transduced by the virus to synthesize the base editor in vivo. Common laboratory vectors of transduction such as lentivirus cause immune responses in humans, so proposed human therapies often centered around adeno-associated virus (AAV) because AAV infections are largely asymptomatic. Unfortunately, the effective packaging capacity of AAV vectors is small, approximately 4.4kb not including inverted terminal repeats.[5] As a comparison, an SpCas9-reverse transcriptase fusion protein is 6.3kb,[6][7] which doesn't even account for the lengthened guide RNA necessary for targeting and priming the site of interest.
See also
References
- Anzalone, Andrew V.; Randolph, Peyton B.; Davis, Jessie R.; Sousa, Alexander A.; Koblan, Luke W.; Levy, Jonathan M.; Chen, Peter J.; Wilson, Christopher; Newby, Gregory A.; Raguram, Aditya; Liu, David R. (21 October 2019). "Search-and-replace genome editing without double-strand breaks or donor DNA". Nature. 576 (7785): 149–157. Bibcode:2019Natur.576..149A. doi:10.1038/s41586-019-1711-4. PMC 6907074. PMID 31634902.
- Ran, F. Ann; Hsu, Patrick D.; Lin, Chie-Yu; Gootenberg, Jonathan S.; Konermann, Silvana; Trevino, Alexandro E.; Scott, David A.; Inoue, Azusa; Matoba, Shogo; Zhang, Yi; Zhang, Feng (September 2013). "Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity". Cell. 154 (6): 1380–1389. doi:10.1016/j.cell.2013.08.021. PMC 3856256. PMID 23992846.
- Sheridan, Cormac (7 November 2019). "Gene editing enters 'prime' time". Nature Biotechnology. doi:10.1038/d41587-019-00032-5.
- "Scientist David Liu takes your questions on CRISPR and prime editing". STAT. 2019-11-06. Retrieved 2020-02-28.
- Wu, Zhijian; Yang, Hongyan; Colosi, Peter (2010). "Effect of Genome Size on AAV Vector Packaging". Molecular Therapy. 18 (1): 80–86. doi:10.1038/mt.2009.255. PMC 2839202. PMID 19904234.
- Anzalone, Andrew V.; Randolph, Peyton B.; Davis, Jessie R.; Sousa, Alexander A.; Koblan, Luke W.; Levy, Jonathan M.; Chen, Peter J.; Wilson, Christopher; Newby, Gregory A.; Raguram, Aditya; Liu, David R. (December 2019). "Search-and-replace genome editing without double-strand breaks or donor DNA". Nature. 576 (7785): 149–157. Bibcode:2019Natur.576..149A. doi:10.1038/s41586-019-1711-4. PMC 6907074. PMID 31634902.
- https://www.addgene.org/132775/
Images created with Biorender.[1]