Haloferax volcanii
Haloferax volcanii is a species of organism in the genus Haloferax in the Archaea.
| Haloferax volcanii | |
|---|---|
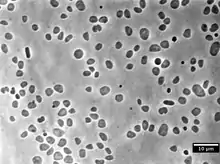 | |
| Haloferax volcanii grown in laboratory conditions and imaged using a phase contrast microscope | |
| Scientific classification | |
| Domain: | |
| Kingdom: | |
| Phylum: | |
| Class: | |
| Order: | |
| Family: | |
| Genus: | |
| Species: | H. volcanii |
| Binomial name | |
| Haloferax volcanii (Mullakhanbhai and Larsen, 1975) Torreblanca et al., 1986 | |
Description and significance
Microbiologist Benjamin Elazari Volcani first discovered Haloferax volcanii, a self-named extremophile, in the 1930s. H. volcanii is a halophilic mesophile archaeon that can be isolated from hypersaline environments such as: the Dead Sea, the Great Salt Lake, and oceanic environments with high sodium chloride concentrates. Haloferax volcanii is noteworthy because it can be cultured without much difficulty, rare for an extremophile. H. volcanii is chemoorganotrophic, metabolizing sugars as a carbon source.[1] It is primarily aerobic, but is capable of anaerobic respiration under anoxic conditions.[2] Recently an isolate of this species was studied by researchers at University of California, Berkeley, as part of a project on the survival of haloarchaea on Mars.
Genome structure
The genome of H. volcanii consists of a large (4 Mb), multicopy chromosome and several megaplasmids. The complete genome, DS2, of H. volcanii consists of about 4130 genes.[3]
The genome has been completely sequenced and a paper discussing it was published in 2010.[4] The molecular biology of H. volcanii has been extensively studied for the last decade in order to discover more about DNA replication, DNA repair and RNA synthesis. The archaeal proteins used in these processes are extremely similar to Eukaryotic proteins and so are studied primarily as a model system for these organisms. H. volcanii undergoes prolific horizontal gene transfer through a mechanism of "mating"- cell fusion.
Cell structure and metabolism
Reproduction among H. volcanii occurs asexually by binary fission. This practice is similar to that of other Archaea and, indeed, that of bacteria.
H. volcanii cells have no cell wall and, like many archaea, therefore use their exterior S-layer for structure. An individual H. volcanii archaeon can vary from 1-3 micrometers in diameter.[5] They are typically recognisable by their 'dished crisp' shape, but are somewhat pleiomorphic so may be seen in other shapes including coccoid.

The membranes of this organism are made of the typical ether linked membrane lipids found solely in archaea and also contain a high level of carotenoids including lycopene, which gives them their distinctive red colour.
H. volcanii use a salt in method to maintain osmostasis, rather than the typical compatible solutes method seen in bacteria. This method involves the maintenance of a high degree of potassium ions in the cell to balance the sodium ions outside. For this reason H. volcanii has a complex ion regulation system and is chemoautotrophic.
H. volcanii will optimally grow at 42 °C in 1.5-2.5 M NaCl and complex nutrient medium. It will still grow at 37 °C, but still requires the concentrated NaCl and complex medium.[3]
Due to the salt in method cytoplasmic proteins are structured to fold in the presence of high ionic concentrations. As such they typically have a large number of charged residues on the exterior section of the protein and very hydrophobic residues forming a core. This structure increases their stability in saline and even high temperature environments considerably, but comes at some loss of processivity compared to bacterial homologs.
H. volcanii respire as their sole source of ATP, unlike several other halobateriacae, such as Halobacterium salinarum they are incapable of photophosphorylation as they lack the necessary bacteriorhodopsin.
Ecology
Isolates of H. volcanii are commonly found in high-salinity aquatic environments, such as the Dead Sea. Their precise role in the ecosystem is uncertain, but the carbohydrates contained within these organisms potentially serve many practical purposes. Because of their ability to maintain homeostasis in spite of the salt around them, H. volcanii could be an important player in advancements in biotechnology. As it is likely that H. volcanii and comparable species are ranked among the earliest living organisms, they also provide information related to genetics and evolution.[6]
Dead Sea
The Dead Sea contains a very high concentration of sodium, magnesium, and calcium salts. This combination makes the sea an ideal environment for extremophiles such as H.volcanii.[7] The Dead Sea has a diverse community of microorganisms, though the field tests completed by Kaplan and Friedman reported that H.volcanii had the largest numerical presence within the community.[8] It is common to find higher numbers of the halophile during the summer, as the Dead Sea is much warmer, averaging around 37 degrees Celsius, and thus more conducive to bacterial blooms.[9] Unfortunately, the Dead Sea is becoming less hospitable to extremophiles such as H. volcanii due to increasing salinity, credited to both natural factors and human activities. As the predominant environment for Haloferax volcanii, the change in salinity places the species at risk.
DNA damage and repair
In prokaryotes the DNA genome is organized in a dynamic structure, the nucleoid, which is embedded in the cytoplasm. Exposure of Haloferax volcanii to stresses that damage the DNA cause compaction and reorganization of the nucleoid.[10] Compaction depends on the Mre11-Rad50 protein complex that is employed in the homologous recombinational repair of DNA double-strand breaks. Delmas et al.[10] proposed that nucleoid compaction is part of a DNA damage response that accelerates cell recovery by helping DNA repair proteins to locate targets, and by facilitating the search for intact DNA sequences during homologous recombination.
Genetic exchange
H. volcanii cells can undergo a pairwise process of genetic exchange which involves cell fusion resulting in a heterodiploid cell (containing two different chromosomes in one cell).[11] Cells of a related species, Haloferax mediterranei, can similarly undergo genetic exchange with each other. H. volcanii has an average nucleotide sequence identity with H. mediterranei of 86.6%. At a reduced frequency, cells of these two species may also interact to undergo genetic exchange.[11] During this process a diploid cell is formed that contains the full genetic repertoire of both parental cells, and genetic recombination is facilitated. Subsequently, the cells separate, giving rise to recombinant cells.
Astrobiology
The conditions Haloferax volcanii survives in, high salinity and high radiation, are very similar to the conditions found on Mars' surface. Consequently, the organism is currently being used to test the survivability of earth native extremophiles on Mars. Advances in this field could lead to a greater understanding of the possibility and time line of extraterrestrial life.[12]
See also
References
- Oren, A. "The Order Halobacteriales." The Prokaryotes: A Handbook on the Biology of Bacteria. 3rd ed. New York: Springer. 2006. pp. 113-164.
- Zaigler, A., Schuster, S.C., and Soppa, J. "Construction and usage of a onefold-coverage shotgun DNA microarray to characterize the metabolism of the archaeon Haloferax volcanii." Molecular Microbiology. 2003. Volume 48, Issue 4, pp. 1089–1105.
- "UCSC Genome Browser Gateway". archaea.ucsc.edu. Retrieved 2017-04-20.
- Hartman, AL; Norais, C; Badger, JH; Delmas, S; Haldenby, S; Madupu, R; Robinson, J; Khouri, H; Ren, Q; Lowe, TM; Maupin-Furlow, J; Pohlschroder, M; Daniels, C; Pfeiffer, F; Allers, T; Eisen, JA (2010). "The complete genome sequence of Haloferax volcanii DS2, a model archaeon". PLoS ONE. 5 (3): e9605. doi:10.1371/journal.pone.0009605. PMC 2841640. PMID 20333302.
- Garrity, G.M., Castenholz, R.W., and Boone, D.R. (Eds.) Bergey's Manual of Systemic Bacteriology, Volume One: The Archaea and the Deeply Branching and Phototrophic Bacteria. 2nd ed. New York: Springer. 2001. p. 316.
- See the NCBI webpage on Haloferax. Data extracted from the "NCBI taxonomy resources". National Center for Biotechnology Information. Retrieved 2007-03-19.
- Oren, A. "Population dynamics of halobacteria in the Dead Sea water column." Limnology and Oceanography. 1983. Volume 28, issue 6, pp. 1094-1103.
- MULLAKIIANBIIAI, M. F., AND H. LARSEN. 1975. Halobacterium volcanii spec. nov., a Dead Sea halobacterium with a modcrate salt requirement. Arch. Mikrobiol. 104: 207-214.
- NEEV, D., ANI) K. 0. EMERY. 1967. The Dead Sea- Depositional processes and environments of evaporites. State of Israel-Min. Develop.-Gcol. Surv. Bull. 41.
- Delmas, S; Duggin, IG; Allers, T (2013). "DNA damage induces nucleoid compaction via the Mre11-Rad50 complex in the archaeon Haloferax volcanii". Mol Microbiol. 87 (1): 168–79. doi:10.1111/mmi.12091. PMC 3565448. PMID 23145964.
- Naor A, Lapierre P, Mevarech M, Papke RT, Gophna U (August 2012). "Low species barriers in halophilic archaea and the formation of recombinant hybrids". Curr. Biol. 22 (15): 1444–8. doi:10.1016/j.cub.2012.05.056. PMID 22748314.
- DasSarma, S. "Extreme Halophiles Are Models for Astrobiology." Microbe Magazine. 2006. Volume 1, No. 3, pp. 120-126.
Further reading
- Carletti, Micaela; Martinez, Maria J.; Gimenez, Maria I.; Sastre, Diego E.; Paggi, Roberto A.; De Castro, Rosana E. (Jun 2014). "The LonB protease controls membrane lipids composition and is essential for viability in the extremophilic haloarchaeon Haloferax volcanii". Environmental Microbiology. 16 (6, Sp. Iss. SI): 1779–1792. doi:10.1111/1462-2920.12385. PMID 24428705. Retrieved 11 November 2014.
- Chimileski, Scott; Franklin, Michael J; Papke, R Thane (14 August 2014). "Biofilms formed by the archaeon Haloferax volcanii exhibit cellular differentiation and social motility, and facilitate horizontal gene transfer". BMC Biology. 12: 65. doi:10.1186/s12915-014-0065-5. PMC 4180959. PMID 25124934.
- Oren A, Ventosa A (2000). "International Committee on Systematic Bacteriology Subcommittee on the taxonomy of Halobacteriaceae. Minutes of the meetings, 16 August 1999, Sydney, Australia". Int. J. Syst. Evol. Microbiol. 50 (3): 1405–1407. doi:10.1099/00207713-50-3-1405. PMID 10843089.
- Parente, Juliana; Casabuono, Adriana; Ferrari, Maria; Paggi, Roberto; De Castro, Rosana; Cuoto, Alicia; Gimenez, Maria (April 18, 2014). "A Rhomboid Protease Gene Deletion Affects a Novel Oligosaccharide N-Linked to the S-layer Glycoprotein of Haloferax volcanii". Journal of Biological Chemistry. 289 (16): 11304–11317. doi:10.1074/jbc.M113.546531. PMC 4036268. PMID 24596091.
- Torreblanca M, Rodriquez-Valera F, Juez G, Ventosa A, Kamekura M, Kates M (1986). "Classification of non-alkaliphilic halobacteria based on numerical taxonomy and polar lipid composition, and description of Haloarcula gen. nov. and Haloferax gen.nov". Syst. Appl. Microbiol. 8 (1–2): 89–99. doi:10.1016/s0723-2020(86)80155-2.
Scientific books
- Gibbons, NE (1974). "Family V. Halobacteriaceae fam. nov.". In RE Buchanan; NE Gibbons (eds.). Bergey's Manual of Determinative Bacteriology (8th ed.). Baltimore: The Williams & Wilkins Co. ISBN 978-0-683-01117-3.