FITM2
Fat storage-inducing transmembrane protein 2 is a protein that in humans is encoded by the FITM2 gene. It plays a role in fat storage. Its location is 20q13.12 and it contains 2 exons.[1] It is also a member of the FIT protein family that has been conserved throughout evolution. Conserved from Saccharomyces cerevisiae to humans is the capability to take fat and store it as cytoplasmic triglyceride droplets.[2] While FIT proteins facilitate the segregation of triglycerides (TGs) into cytosolic lipid droplets, they are not involved in triglyceride biosynthesis.[3] In mammals, both FIT2 and FIT1 from the same family are present, embedded in the wall of the endoplasmic reticulum (ER) where they regulate lipid droplet formation in the cytosol. In S. cerevisiae, it also plays a role in the metabolism of phospholipids.[4] These TGs are in the cytoplasm, encapsulated by a phospholipid monolayer in configurations or organelles that have been given many different names including lipid particles, oil bodies, adiposomes, eicosasomes, and most prevalent in scientific research – lipid droplets.[2]
FIT protein family
FITM2 one of two genes in its family. The other being FITM1 also known as FIT1 in which it shares 35% identity with.[4] However, FITM1 and FITM2 have a similarity score of 50% at the amino acid level. Of the two protein coding genes, FITM2 is the ancient orthologue of this family of FIT proteins with orthologues also found in S.cerevisiae.[3] FITM1 is also found in humans but is conserved from fish. FITM1 is not seen in adipose tissue or adipocytes but it is however, displayed mostly in muscles both skeletal and cardiac in nature.[3] FITM2 is seen most frequently and in increased expression in adipose tissue. It is controlled by receptor γ (peroxisome proliferator activated) directly. This receptor γ is the principal transcription factor for the differentiation of adipocytes.[4]
Lipid droplets (LDs)
Cytosolic lipid droplets are organelles that are composed of a core that is hydrophobic in nature containing neutral lipids (like triglycerides) as well as cholesteryl esters that have a phospholipid monolayer in addition to a distinctive set of expressed proteins that surrounds them.[4] The most generally accepted view on the creation of lipid droplets is that the neutral lipids build up between the ER leaflets due to de novo synthesizing enzymes for both triglyceride phospholipids and cholesteryl esters. This leads to the budding lipid droplets growing into the cytoplasm space.[4] There are two different groups of lipid droplets that are known: the first is characterized by its phospholipid leaflet in continuity with the membrane of the ER and the second is classified as definitively cytosolic without a connection to the ER.[4]
Structure
A generally acknowledged model of the creation of a lipid droplet includes the construction of a center or lens of TGs that are produced new. This TG center is flanked by the leaflets of the membrane in the ER that sprouts off with the leaflet in the cytoplasm of the ER that surrounds the core of the lipid (neutral). It is then able to obtain interchangeable proteins that are associated with lipid droplets in the cytosol.[2]
Studies done have suggested that FITM2 works downstream of diglyceride acyltransferase (DGAT) enzymes and binds to TGs, which is crucial for a cell’s FITM2 facilitated lipid droplet formation after being purified.[8] When looking at the most recent view of lipid droplet formation as described above where a TG lens is established between ER leaflets, FITM2’s capacity to bind TG may aid in the increase of TG’s solubility in the ER. This can then instigate the gathering of amounts of TG necessary to mediate the progression of lipid droplet formation. Consequently, FITM2 has been referred to as a “gatekeeper” because it is situated downstream of TG biosynthesis and controls the number of lipid droplets formed.[8]
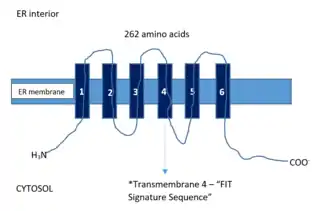
In mammals, FITM2 protein is made up of 262 amino acids (while FIT1 part of the same family is 292 amino acids long) and has six transmembrane domains in which the N and C termini are both geared to face the cytosol.[3] When FITM2 has a mutation in its fourth transmembrane domain that happens to be a gain-of-function one, is found overexpressed in cells, it has unfailingly caused the buildup of TG rich lipid droplets. This mutation has been described as having a significant effect on increasing both the amount and size and lipid droplets.[4] A comparative sequence analysis of FITM2 showed a tract of residues that were deemed as extensively conserved located in this transmembrane 4 that was later named the “FIT signature sequence”.[9]
Function
In the cells of mammals, the construction of lipid droplets is a process that is strictly controlled, using hormone induced signals, proteins related to droplets, and lipases as well. Four observations support the role of FIT proteins in the buildup and mediation of lipid droplets.[2] First, they have been conserved throughout evolution and solely found in the ER which is the primary site for the biosynthesis of TGs.[2] Second, when FIT proteins are overexpressed in either the liver of a mouse of even in cells that have been cultured in vivo, there has been observable buildup of lipid droplets that are rich in triglycerides as an outcome.[2] Third, FIT proteins are not DGATs. DGATs facilitate the biosynthesis of the TGs. FIT proteins strictly aid in the conversion of the TGs (made by DGATs) into lipid droplets. Therefore, knowing the function of these FIT proteins helps us to make sense of why they are placed downstream of the DGATs.[2] Lastly, a shRNA-facilitated reduction in FITM2 in adipocytes (3T3-L1) or even a knockdown of it in the embryos of zebrafish resulted in great declines in lipid droplet build-up.[2]
FITM2 has been identified as being overexpressed throughout the time 3T3-L1 (from the adipocyte cell line) is being differentiated which shows resemblance to the peroxisome proliferator-activated receptor gamma (PPAR γ) at a specific period when lipid droplets have been identified to build-up which results in the adipocyte phenotype that is seen in the 3T3-L1 cells.[2] The overexpression of FITM2 was also displayed when the 3T3-L1 cells were combined with rosiglitazone (a PPAR γ agonist). This serves as evidence for the idea FITM2 is functionally regulated by PPAR γ.[2]
The specificity of the tissue dispersal of FITM1 and FITM2 and the fact that FITM2 binds TG more intensely than FITM1 (which forms a weak bond) presents separate functions for these FIT family proteins in regards to the metabolism of lipids.[3] Lipid droplet development induced by FITM2 may function in TG storage for long-term purposes in adipose whereas FITM1 may function to make the smaller lipid droplets that are seen in skeletal muscle where there is a fast replacement of LDs.[3]
Clinical Uses
When physiological circumstances are regular, lipid droplets are depended on to keep energy balanced at not only at the cellular level, but for the sake of the whole organism’s sustainability. However, excessive acquirement of lipid droplets can result in obesity, and increased risk for obtaining disease including type 2 diabetes, atherosclerosis, and heart disease.[2] The documentation of the FIT proteins should help in evolving substances to revert FIT expression or activity back to a normal regulatory state as treatment for these diseases.
In addition, a recent study was performed on a family with a new homozygous mutation in FITM2, resulting in a truncated protein. The individuals in the family that are affected by this mutation exhibit Siddiqi syndrome. Siddiqi syndrome is identified by gradual development of hearing loss, late motor development, decreased BMI, ichthyosis-like changes to the skin, and minor fiber neuropathy.[10] In this family, the collection of symptoms presented for this syndrome is new. However, they also overlap with several recognized monogenic conditions that are neurological in nature including Troyer syndrome, Mohr-Tranebjaerg syndrome, and Megdel syndrome.[10]
References
- "FITM2 fat storage inducing transmembrane protein 2 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2017-11-29.
- Kadereit B, Kumar P, Wang WJ, Miranda D, Snapp EL, Severina N, Torregroza I, Evans T, Silver DL (January 2008). "Evolutionarily conserved gene family important for fat storage". Proceedings of the National Academy of Sciences of the United States of America. 105 (1): 94–9. Bibcode:2008PNAS..105...94K. doi:10.1073/pnas.0708579105. PMC 2224239. PMID 18160536.
- Gross DA, Zhan C, Silver DL (December 2011). "Direct binding of triglyceride to fat storage-inducing transmembrane proteins 1 and 2 is important for lipid droplet formation". Proceedings of the National Academy of Sciences of the United States of America. 108 (49): 19581–6. doi:10.1073/pnas.1110817108. PMC 3241795. PMID 22106267.
- Goh VJ, Tan JS, Tan BC, Seow C, Ong WY, Lim YC, Sun L, Ghosh S, Silver DL (October 2015). "Postnatal Deletion of Fat Storage-inducing Transmembrane Protein 2 (FIT2/FITM2) Causes Lethal Enteropathy". The Journal of Biological Chemistry. 290 (42): 25686–99. doi:10.1074/jbc.M115.676700. PMC 4646211. PMID 26304121.
- GRCh38: Ensembl release 89: ENSG00000197296 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Miranda DA, Kim JH, Nguyen LN, Cheng W, Tan BC, Goh VJ, Tan JS, Yaligar J, Kn BP, Velan SS, Wang H, Silver DL (April 2014). "Fat storage-inducing transmembrane protein 2 is required for normal fat storage in adipose tissue". The Journal of Biological Chemistry. 289 (14): 9560–72. doi:10.1074/jbc.M114.547687. PMC 3975007. PMID 24519944.
- Gross DA, Snapp EL, Silver DL (May 2010). "Structural insights into triglyceride storage mediated by fat storage-inducing transmembrane (FIT) protein 2". PLOS ONE. 5 (5): e10796. doi:10.1371/journal.pone.0010796. PMC 2875400. PMID 20520733.
- Zazo Seco C, Castells-Nobau A, Joo SH, Schraders M, Foo JN, van der Voet M, et al. (February 2017). "A homozygous FITM2 mutation causes a deafness-dystonia syndrome with motor regression and signs of ichthyosis and sensory neuropathy". Disease Models & Mechanisms. 10 (2): 105–118. doi:10.1242/dmm.026476. PMC 5312003. PMID 28067622.
Further reading
- Cho YS, Chen CH, Hu C, Long J, Ong RT, Sim X, et al. (December 2011). "Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians". Nature Genetics. 44 (1): 67–72. doi:10.1038/ng.1019. PMC 3582398. PMID 22158537.
- Kadereit B, Kumar P, Wang WJ, Miranda D, Snapp EL, Severina N, Torregroza I, Evans T, Silver DL (January 2008). "Evolutionarily conserved gene family important for fat storage". Proceedings of the National Academy of Sciences of the United States of America. 105 (1): 94–9. Bibcode:2008PNAS..105...94K. doi:10.1073/pnas.0708579105. PMC 2224239. PMID 18160536.
This article incorporates text from the United States National Library of Medicine, which is in the public domain.

