CrfA RNA
CrfA RNA (Caulobacter response to famine RNA) is a family of non-coding RNAs found in Caulobacter crescentus. CrfA is expressed upon carbon starvation and is thought to activate 27 genes.[1] It was originally identified along with 26 other non-coding RNAs using a tiled Caulobacter microarray protocol specifically aimed at detecting small RNAs.[2]
| CrfA RNA | |
|---|---|
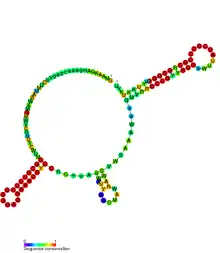 Conserved secondary structure of CrfA RNA. | |
| Identifiers | |
| Symbol | CrfA |
| Other data | |
| Domain(s) | Caulobacter |
| PDB structures | PDBe |
CrfA RNA is one of only 8 putative ncRNAs conserved in the closely related Caulobacter sp. K31.[1]
Carbon starvation response
CrfA was found to be upregulated 10-fold in C. crescentus when glucose was deprived in a minimal medium. Further experimentation found the response to be specific to carbon deprivation; its expression was not increased when phosphate or nitrogen were limiting factors.[1]
Affymetrix microarrays were then used to analyse changes in the transcriptome in response to increased CrfA. Seven affected gene products were TonB-dependent receptors, outer membrane proteins which facilitate the uptake of external substrate.[3] The upregulation of these proteins could increase the carbon uptake during starvation. Another CrfA regulated gene (EntrezGene CC_1363) is thought to encode a proton pump powered by pyrophosphate hydrolysis. Increasing production of this protein could enable the cell to maintain its electrochemical gradient and power ATP synthesis during carbon starvation.[1]
σ54 regulates many other proteins involved in the carbon starvation response at the level of transcription.[4]
References
- Landt SG, Lesley JA, Britos L, Shapiro L (September 2010). "CrfA, a Small Noncoding RNA Regulator of Adaptation to Carbon Starvation in Caulobacter crescentus". J. Bacteriol. 192 (18): 4763–4775. doi:10.1128/JB.00343-10. PMC 2937403. PMID 20601471.
- Landt SG, Abeliuk E, McGrath PT, Lesley JA, McAdams HH, Shapiro L (May 2008). "Small non-coding RNAs in Caulobacter crescentus". Mol. Microbiol. 68 (3): 600–614. doi:10.1111/j.1365-2958.2008.06172.x. PMID 18373523.
- Koebnik R (August 2005). "TonB-dependent trans-envelope signalling: the exception or the rule?". Trends Microbiol. 13 (8): 343–347. doi:10.1016/j.tim.2005.06.005. PMID 15993072.
- England JC, Perchuk BS, Laub MT, Gober JW (February 2010). "Global regulation of gene expression and cell differentiation in Caulobacter crescentus in response to nutrient availability". J. Bacteriol. 192 (3): 819–833. doi:10.1128/JB.01240-09. PMC 2812448. PMID 19948804.
Further reading
- Laub MT, Shapiro L, McAdams HH (2007). "Systems biology of Caulobacter" (PDF). Annu. Rev. Genet. 41: 429–441. doi:10.1146/annurev.genet.41.110306.130346. PMID 18076330.
- Hinz AJ, Larson DE, Smith CS, Brun YV (February 2003). "The Caulobacter crescentus polar organelle development protein PodJ is differentially localized and is required for polar targeting of the PleC development regulator". Mol. Microbiol. 47 (4): 929–941. doi:10.1046/j.1365-2958.2003.03349.x. PMID 12581350.
- Massé E, Majdalani N, Gottesman S (April 2003). "Regulatory roles for small RNAs in bacteria". Curr. Opin. Microbiol. 6 (2): 120–124. doi:10.1016/S1369-5274(03)00027-4. PMID 12732300.
- Majdalani N, Vanderpool CK, Gottesman S (2005). "Bacterial small RNA regulators". Crit. Rev. Biochem. Mol. Biol. 40 (2): 93–113. doi:10.1080/10409230590918702. PMID 15814430.
- Ely B (1991). "Genetics of Caulobacter crescentus". Meth. Enzymol. 204: 372–384. doi:10.1016/0076-6879(91)04019-K. PMID 1658564.
- Fröhlich KS, Vogel J (December 2009). "Activation of gene expression by small RNA". Curr. Opin. Microbiol. 12 (6): 674–682. doi:10.1016/j.mib.2009.09.009. PMID 19880344.