Betabaculovirus
Betabaculovirus is a genus of viruses, in the family Baculoviridae. Arthropods serve as natural hosts. There are currently 26 species in this genus including the type species Cydia pomonella granulovirus.[1][2]
| Betabaculovirus | |
|---|---|
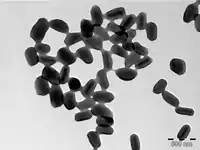 | |
| Electron micrograph of Phthorimaea operculella granulovirus occlusion bodies | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | incertae sedis |
| Kingdom: | incertae sedis |
| Phylum: | incertae sedis |
| Class: | incertae sedis |
| Order: | incertae sedis |
| Family: | Baculoviridae |
| Genus: | Betabaculovirus |
| Type species | |
| Cydia pomonella granulovirus | |
Taxonomy
Group: dsDNA
- Family: Baculoviridae
- Genus: Betabaculovirus
- Adoxophyes orana granulovirus
- Agrotis segetum granulovirus
- Artogeia rapae granulovirus
- Choristoneura fumiferana granulovirus
- Clostera anachoreta granulovirus
- Clostera anastomosis granulovirus A
- Clostera anastomosis granulovirus B
- Cnaphalocrocis medinalis granulovirus
- Cryptophlebia leucotreta granulovirus
- Cydia pomonella granulovirus
- Diatraea saccharalis granulovirus
- Epinotia aporema granulovirus
- Erinnyis ello granulovirus
- Harrisina brillians granulovirus
- Helicoverpa armigera granulovirus
- Lacanobia oleracea granulovirus
- Mocis latipes granulovirus
- Mythimna unipuncta granulovirus A
- Mythimna unipuncta granulovirus B
- Phthorimaea operculella granulovirus
- Plodia interpunctella granulovirus
- Plutella xylostella granulovirus
- Spodoptera frugiperda granulovirus
- Spodoptera litura granulovirus
- Trichoplusia ni granulovirus
- Xestia c-nigrum granulovirus
Structure
Viruses in Betabaculovirus are enveloped. Genomes are circular, around 80-180kb in length. The genome codes for 100 to 180 proteins.[2]
| Genus | Structure | Symmetry | Capsid | Genomic arrangement | Genomic segmentation |
|---|---|---|---|---|---|
| Betabaculovirus | Budded or Occluded | Enveloped | Circular | Monopartite |
Life cycle
Viral replication is nuclear. Entry into the host cell is achieved by attachment of the viral glycoproteins to host receptors, which mediates endocytosis. Replication follows the dsDNA bidirectional replication model. Dna templated transcription, with some alternative splicing mechanism is the method of transcription. The virus exits the host cell by nuclear pore export, and existing in occlusion bodies after cell death and remaining infectious until finding another host. Arthropods serve as the natural host. Transmission routes are fecal-oral.[2]
| Genus | Host details | Tissue tropism | Entry details | Release details | Replication site | Assembly site | Transmission |
|---|---|---|---|---|---|---|---|
| Betabaculovirus | Arthropods | Midgut then hemocoel; digestive gland epithelium (shrimps) | Cell receptor endocytosis | Budding; Occlusion | Nucleus | Nucleus | Oral-fecal |
References
- Harrison, RL; Herniou, EA; Jehle, JA; Theilmann, DA; Burand, JP; Becnel, JJ; Krell, PJ; van Oers, MM; Mowery, JD; Bauchan, GR; Ictv Report, Consortium (September 2018). "ICTV Virus Taxonomy Profile: Baculoviridae". The Journal of General Virology. 99 (9): 1185–1186. doi:10.1099/jgv.0.001107. PMID 29947603.
- "Viral Zone". ExPASy. Retrieved 15 June 2015.