Anti small RNA
Antisense small RNA are short RNA sequences (about 50-500 nucleotides long) that are complementary to other small RNA (sRNA) in the cell. [2]
| Anti GcvB sRNA | |
|---|---|
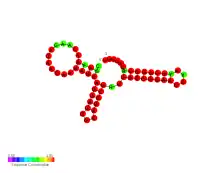 Predicted secondary structure and sequence conservation Anti GcvB sRNA | |
| Identifiers | |
| Rfam | RF02702 |
| Other data | |
| Domain(s) | Bacteria |
| GO | GO:0045975 |
| SO | SO:0000370 |
| PDB structures | PDBe |
| Anti stx2 sRNA | |
|---|---|
 Predicted secondary structure and sequence conservation Anti stx2 sRNA | |
| Identifiers | |
| Rfam | RF02703 |
| Other data | |
| Domain(s) | Bacteria |
| GO | GO:0045975 |
| SO | SO:0000370 |
| PDB structures | PDBe |
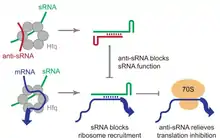
sRNAs can repress translation via complementary base-pairing with their target mRNA sequence.[3] Anti-sRNAs function by complementary pairing with sRNAs before the mRNA can be bound, thus freeing the mRNA and relieving translation inhibition.[1]
Function
Antisense small RNA are found in all of life including Eukaryotes, Bacteria and Archaea.[4][5] They are non-coding RNA sequences involved in regulatory processes, metabolism, and aiding transcription.[4]
Antisense RNA can also be engineered and utilized by scientists to perform experimental functions.[6]
Identification Methods
Numerous studies have been performed to identify potential antisense sRNA candidates. Recent experiments have used Northern blot analysis and 5'-end mapping to correctly identify potential antisense sRNA candidates.[7] In 2019, a new algorithm called APERO was established which allows the detection of small transcripts from paired-end bacterial RNA-seq data. [8] RNA-seq is a popular method used for the identification of small RNA. [8] However, while reliable for eukaryotic sRNA, it remains inaccurate for bacterial sRNA. [8]
Examples
AsxR
AsxR, previously known as EcOnc02, is an anti-sRNA encoded within the 3' region of the stx2B gene of E.Coli bacteria.[1] It acts to increase expression of the ChuS heme oxygenase via destabilisation of FnrS sRNA.[1] This aids bacterial infection of the animal host gut. [1]
References
- Tree JJ, Granneman S, McAteer SP, Tollervey D, Gally DL (July 2014). "Identification of bacteriophage-encoded anti-sRNAs in pathogenic Escherichia coli". Molecular Cell. 55 (2): 199–213. doi:10.1016/j.molcel.2014.05.006. PMC 4104026. PMID 24910100.
- Bhatt S, Egan M, Jenkins V, Muche S, El-Fenej J (2016). "Escherichia coli". Frontiers in Cellular and Infection Microbiology. 6: 105. doi:10.3389/fcimb.2016.00105. PMC 5030294. PMID 27709103.
- Grosshans H, Filipowicz W (January 2008). "Molecular biology: the expanding world of small RNAs". Nature. 451 (7177): 414–6. Bibcode:2008Natur.451..414G. doi:10.1038/451414a. PMID 18216846.
- Bernick DL, Dennis PP, Lui LM, Lowe TM (2012). "Diversity of Antisense and Other Non-Coding RNAs in Archaea Revealed by Comparative Small RNA Sequencing in Four Pyrobaculum Species". Frontiers in Microbiology. 3: 231. doi:10.3389/fmicb.2012.00231. PMC 3388794. PMID 22783241.
- Figueroa-Bossi N, Valentini M, Malleret L, Fiorini F, Bossi L (September 2009). "Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target". Genes & Development. 23 (17): 2004–15. doi:10.1101/gad.541609. PMC 2751969. PMID 19638370.
- Rodrigo G, Prakash S, Cordero T, Kushwaha M, Jaramillo A (February 2016). "Functionalization of an Antisense Small RNA". Journal of Molecular Biology. Engineering Tools and Prospects in Synthetic Biology. 428 (5 Pt B): 889–92. doi:10.1016/j.jmb.2015.12.022. PMC 4819895. PMID 26756967.
- Thomason MK, Storz G (2010). "Bacterial antisense RNAs: how many are there, and what are they doing?". Annual Review of Genetics. 44 (1): 167–88. doi:10.1146/annurev-genet-102209-163523. PMC 3030471. PMID 20707673.
- Leonard S, Meyer S, Lacour S, Nasser W, Hommais F, Reverchon S (September 2019). "APERO: a genome-wide approach for identifying bacterial small RNAs from RNA-Seq data". Nucleic Acids Research. 47 (15): e88. doi:10.1093/nar/gkz485. PMC 6735904. PMID 31147705.