AT-hook
The AT-hook is a DNA-binding motif present in many proteins, including the high mobility group (HMG) proteins,[1] DNA-binding proteins from plants [2] and hBRG1 protein, a central ATPase of the human switching/sucrose non-fermenting (SWI/SNF) remodeling complex.[3]
| AT-hook | |||||||||
|---|---|---|---|---|---|---|---|---|---|
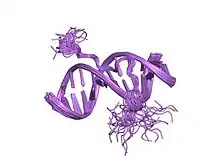 solution structure of a complex of the second dna binding domain of human hmg-i(y) bound to dna dodecamer containing the prdii site of the interferon-beta promoter, nmr, 35 structures | |||||||||
| Identifiers | |||||||||
| Symbol | AT_hook | ||||||||
| Pfam | PF02178 | ||||||||
| InterPro | IPR017956 | ||||||||
| SMART | AT_hook | ||||||||
| SCOP2 | 2eze / SCOPe / SUPFAM | ||||||||
| |||||||||
This motif consists of a conserved, palindromic, core sequence of proline-arginine-glycine-arginine-proline, although some AT-hooks contain only a single proline in the core sequence. AT-hooks also include a variable number of positively charged lysine and arginine residues on either side of the core sequence.[4] The AT-hook binds to the minor groove of adenine-thymine (AT) rich DNA, hence the AT in the name. The rest of the name derives from a predicted asparagine/aspartate "hook" in the earliest AT-hooks reported in 1990.[5] In 1997 structural studies using NMR determined that a DNA-bound AT-hook adopted a crescent or hook shape around the minor groove of a target DNA strand (pictured at right).[6] HMGA proteins contain three AT-hooks, although some proteins contain as many as 30.[5] The optimal binding sequences for AT-hook proteins are repeats of the form (ATAA)n or (TATT)n, although the optimal binding sequences for the core sequence of the AT-hook are AAAT and AATT.[7]
References
- Reeves R, Beckerbauer L (May 2001). "HMGI/Y proteins: flexible regulators of transcription and chromatin structure". Biochim. Biophys. Acta. 1519 (1–2): 13–29. doi:10.1016/S0167-4781(01)00215-9. PMID 11406267.
- Meijer AH, van Dijk EL, Hoge JH (June 1996). "Novel members of a family of AT hook-containing DNA-binding proteins from rice are identified through their in vitro interaction with consensus target sites of plant and animal homeodomain proteins". Plant Mol. Biol. 31 (3): 607–18. doi:10.1007/BF00042233. PMID 8790293. S2CID 24687309.
- Singh M, D'Silva L, Holak TA (2006). "DNA-binding properties of the recombinant high-mobility-group-like AT-hook-containing region from human BRG1 protein". Biol. Chem. 387 (10–11): 1469–78. doi:10.1515/BC.2006.184. PMID 17081121.
- Reeves R (October 2001). "Molecular biology of HMGA proteins: hubs of nuclear function". Gene. 277 (1–2): 63–81. doi:10.1016/S0378-1119(01)00689-8. PMID 11602345.
- Reeves R, Nissen MS (May 1990). "The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure". J. Biol. Chem. 265 (15): 8573–82. PMID 1692833.
- Huth JR, Bewley CA, Nissen MS, et al. (August 1997). "The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif". Nat. Struct. Biol. 4 (8): 657–65. doi:10.1038/nsb0897-657. PMID 9253416.
- Reeves R (October 2000). "Structure and function of the HMGI(Y) family of architectural transcription factors". Environ. Health Perspect. 108 (Suppl 5): 803–9. doi:10.2307/3454310. JSTOR 3454310. PMID 11035986. Archived from the original on 2009-01-09.